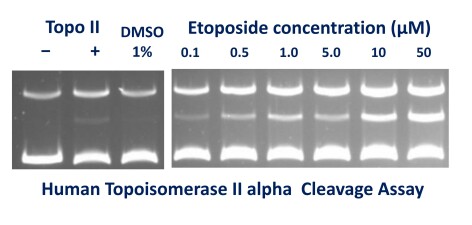
Cleavage Assays
Interruption of the DNA breakage-reunion step or stabilisation of the DNA-breakage step of the topoisomerase reaction can lead to cell death and is the mechanism by which many successful topoisomerase inhibitors work (e.g. the quinolone antibacterials and the anticancer drug etoposide). Cleavage assays are particularly useful in determining if a potential compound acts by this mechanism. Quinolones and similar compounds do not require ATP to stabilise the cleavage intermediate, thus the cleavage assay buffer does not contain ATP. However, if you are concerned that your compound/molecule of interest requires ATP to stabilise the cleaved intermediate, ATP can be added to the reaction.
Although all topoisomerases cleave DNA, cleavage assays can occasionally problematic. They often require more enzyme than required in relaxation/supercoiling or decatenation, and, as we have often found, require high quality enzymes with high specific activity. For example you require almost 10x more E. coli DNA gyrase for cleavage reactions than you do for supercoiling reactions.
A typical reaction:
Enzyme is incubated with 0.5 µg of supercoiled pBR322 in a reaction volume of 30 µl at 37°C for 30 minutes in 1X Cleavage Assay Buffer (with or without ATP as required). 0.2% (w/v) SDS and 0.1 mg/ml Proteinase K are added before a further incubation at 37°C for 30 minutes.
The reaction is stopped by the addition of 3 µl 0.5 mM EDTA and 30 µl chloroform/iso-amyl alcohol (24/1) and 30 µl Stop Buffer (STEB), before being loaded on an agarose gel.
Gels can be run either in TBE (90 mM Tris.Borate, 2 mM EDTA) or TAE (40 mM Tris.acetate, 2 mM EDTA) with or without an intercalator such as ethidium bromide.
Typical Gel: