Linking number and the Unwinding Assays
The DNA Intercalation Assay
This assay is designed to investigate whether a compound intercalates into the DNA double-helix, or binds in the minor groove (note 1), leading to unwinding of the DNA. This is a characteristic of a number of inhibitors of DNA-modifying enzymes, such as the topoisomerase inhibitor amsacrine (m-AMSA).
The Linking Number (Lk) of DNA
The linking number of DNA defines the number of times a strand of DNA winds around the helical axis when the axis is constrained to lie in a plane. If both strands are covalently intact, the linking number cannot change. For instance, for a circular, relaxed DNA such as pBR322 of 4361 base pairs, the linking number is 4361/10.5= +415, where 10.5 is the approximate number of base pairs per turn for B-form DNA (note 2).
Lk = Tw+Wr
Where Tw (twist) is a measure of the helical winding of the DNA strands around each other,
and Wr (writhe) is a measure of the coiling of the axis of the double helix. A right-handed coil is assigned a negative number (negative supercoiling) and a left-handed coil is assigned a positive number (positive supercoiling).
The twist and writhe of any given DNA molecule must sum to the linking number: Lk = Tw + Wr, so any change in the twist of the DNA will result in an equal and opposite change in the writhe (if the linking number is fixed) and any change in linking number results in a change in the twist and/or writhe
(ΔLk = ΔTw + ΔWr).
If there is no supercoiling (i.e. relaxed DNA), then Wr = 0 and Tw = Lk (= +415 for pBR322).
The example below shows how twist and writhe can vary while linking number remains constant for a DNA circle, i.e. depending upon solution conditions etc., ΔLk can be partitioned between ΔTw and ΔWr.
The example below shows how twist and writhe can vary while linking number remains constant for a DNA circle, i.e. depending upon solution conditions etc., ΔLk can be partitioned between ΔTw and ΔWr.
|
|
Relaxed |
Positively supercoiled |
Negatively supercoiled |
||||||
|
ΔLk |
0 |
+3 |
+3 |
+3 |
+3 |
-3 |
-3 |
-3 |
-3 |
|
ΔTw |
0 |
+3 |
+2 |
+1 |
0 |
-3 |
-2 |
-1 |
0 |
|
ΔWr |
0 |
0 |
+1 |
+2 |
+3 |
0 |
-1 |
-2 |
-3 |
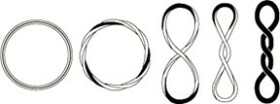 |
 |
The Intercalation Assay
The relationship between twist, writhe and linking number is the basis of this assay. Intercalators contain planar, normally polycyclic, aromatic structures which can insert between the bases of the double-helical DNA molecule. Compounds which are able to intercalate into DNA or bind in the groove can lead to local unwinding of the DNA leading to a decrease in the twist of the DNA. Since the linking number is fixed (the strands remain intact) then there must be an increase in writhe to compensate (i.e. the DNA will become positively supercoiled).
If the DNA is then incubated with a topoisomerase such as wheatgerm topo I it will remove these supercoils and relax the DNA so that the writhe becomes zero. On removal of the compound (by butanol extraction) the twist will increase. Since the linking number is now fixed (the butanol extraction removes the enzyme as well) the increase in twist must be compensated by a decrease in writhe. This leads to a more negatively supercoiled plasmid which has a higher mobility and runs faster on an agarose gel (note 3). The supercoiled DNA formed in these conditions is indicative of an intercalator (note 4).
This is the basis of the DNA unwinding assay. Relaxed (or supercoiled) plasmid DNA is incubated briefly with the test compound prior to relaxation by the wheat germ topo I. The enzyme and drug are then removed by butanol extraction and the plasmid analysed by gel electrophoresis. The change in the degree of supercoiling indicates if the compound is an intercalator. Performing the same experiment with relaxed and supercoiled plasmid eliminates potential false negatives and positives. The table below outlines potential outcomes of the intercalation assay and their interpretation.
Possible results of the assay
|
|
Substrate |
Compound |
|
a |
Relaxed DNA |
Compound is an intercalator but not a topo I inhibitor |
|
The intercalation leads to a decreased twist and a consequent increased writhe (positive supercoils). The topo I relaxes the DNA so writhe becomes zero. Removal of compound and enzyme increases twist so writhe decreases giving negatively supercoiled DNA. |
||
|
Result: Negatively supercoiled DNA |
||
|
|
||
|
b |
Relaxed DNA |
Compound is neither an intercalator nor a topo I inhibitor |
|
There is no change in twist or writhe. The DNA remains relaxed so the enzyme has no affect. |
||
|
Result: Relaxed DNA |
||
|
|
|
|
|
c |
Relaxed DNA |
Compound is both an intercalator and a topo I inhibitor |
|
The intercalation leads to a decreased twist and a consequent increased writhe (positive supercoils). The topo I is inhibited so does not relax the DNA. Removal of compound and enzyme reverses the original change in twist so the plasmid runs as relaxed DNA. It is a false negative since the result is as #b. |
||
|
Result: Relaxed DNA |
||
|
|
||
|
d |
Negatively supercoiled |
Compound is an intercalator but not a topo I inhibitor |
|
The intercalation leads to decreased twist and a consequent increased writhe (the DNA may become less negatively supercoiled, relaxed or positively supercoiled depending on the potency of the intercalator). The topo I relaxes the DNA (if negatively or positively supercoiled) so writhe becomes zero. Removal of compound (and enzyme) increases twist so writhe decreases giving negatively supercoiled DNA. Depending on the degree of supercoiling this will run as a ladder of topoisomers on the gel (for poor intercalators or low concentrations of strong intercalators) or at the same point as the substrate (for strong intercalators). |
||
|
Result: Negatively supercoiled DNA |
||
|
|
|
|
|
e |
Negatively supercoiled |
Compound is neither an intercalator nor a topo I inhibitor |
|
There is no change in twist. The DNA is relaxed by the enzyme and runs as such on the gel |
||
|
Result: Relaxed DNA |
||
|
|
|
|
|
f |
Negatively supercoiled |
Compound is both an intercalator and a topo I inhibitor |
|
The intercalation leads to a decreased twist and a consequent increased writhe (positive supercoils). The topo I is inhibited so does not relax the DNA. Removal of compound and enzyme reverses the original change in twist so the plasmid runs as negatively supercoiled DNA. It is a false positive since the result can run as #d |
||
|
Result: Negatively supercoiled DNA |
||
Notes
1. Minor groove binders such as netropsin can increase the twist and therefore decrease the writhe of the DNA, the opposite of the intercalators described.
2. More accurately, this is the hypothetical linking number Lk0 under standard conditions, which in this case is +415.333. However, the actual linking number Lk has to be an integer, since the ends of the strands must align to be connected, so Lk = +415 for this relaxed plasmid. Under standard conditions for relaxed DNA, Lk is called Lkm. If the DNA is supercoiled then the linking number Lk will change. The linking difference (ΔLk) is Lk-Lkm. So, for example, pBR322 (Lkm = +415) with a linking number of +412 has a linking difference (ΔLk) of -3. This is underwound or negatively supercoiled DNA. (For a more detailed description see: Bates, A. D. & Maxwell, A. (2005). DNA Topology, Oxford University Press, Oxford.)
3. This applies to agarose gels run in the absence of any intercalators (e.g. chloroquine or ethidium bromide). If they are included in the gel then these will change the way that the topoisomers migrate. They will intercalate into the DNA causing a decrease in twist and therefore an increase in writhe. An agarose gel run in the absence of an intercalator is not able to resolve all the topoisomers, usually only up to a linking difference of about 12 to 14. For example, negatively supercoiled pBR322 DNA with linking differences of -18 to -22 will run as a single band. If these samples are run in the presence of low chloroquine concentrations then they will become more relaxed (twist decreases so writhe increases, effectively a decrease in negative writhe) allowing them to be resolved as a series of topoisomers. If the concentration of chloroquine is increased they will become relaxed and at even higher concentrations they will become positively supercoiled. Note however that the linking number has not changed, just the degree of writhe.
4. If the topo I was not used at all then the linking number would be constant. If relaxed plasmid was incubated with the compound this would decrease the twist leading to an increase in writhe (it would become positively supercoiled). It might be thought that this could just be run on a gel to see the difference. However, the intercalator is likely to dissociate from the DNA during the electrophoresis so the plasmid would revert to being relaxed and no change could be seen. The use of the topo I changes the linking number while the intercalator is present. Once the enzyme has been removed the linking number cannot change further and so the effect of the intercalator is made ‘permanent’.
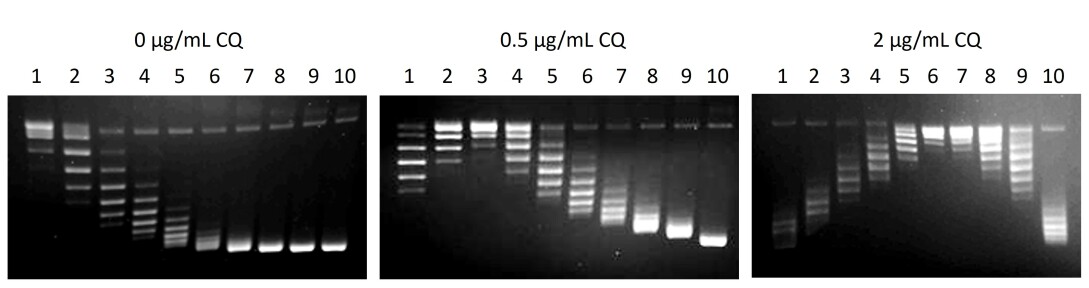
Analysis of pBR322 with different linking numbers on agarose gels
2 µl of each marker was loaded onto a 1% agarose gel and run in TAE buffer. Three gels are shown run in the presence of 0, 0.5 and 2 µg/mL chloroquine (CQ) to resolve the different topoisomers. Using the central topoisomer of sample 1 (relaxed pBR322) as a linking number difference of 0 (Lkm), it is possible to use the different markers to count the linking numbers up to about -23 (negative supercoiling).