ATPase Reactions
Apart from the ATP-independent relaxation of supercoiled DNA by DNA gyrase, all type II topoisomerases and the type I topoisomerase, Reverse Gyrase, require ATP to catalyse the interconversion between the different topological isoforms of DNA.
Binding and hydrolysis of ATP and the release of ADP and Pi are transduced through the enzyme and happen alongside important conformational changes that drive the topoisomerase reaction cycle.
The ATP binding site (GHKL domain) is a target for drugs. In particular, the coumarin drugs bind to the N-terminal domain of GyrB/ParE competitively inhibiting the hydrolysis of ATP. ATPase assay can be used to find compounds that inhibit this part of the reaction cycle. The ATPase assay that we supply is a continuous coupled assay which can provide more information (such as accurate kinetic information) unlike endpoint assays such as the Malachite green ATPase assay.
The coupled enzyme ATPase assay is based on the conversion of phosphoenolpyruvate (PEP) to pyruvate by pyruvate kinase (PK) coupled to the conversion of pyruvate to lactate by lactate dehydrogenase (LDH). This step requires NADH which is oxidized to NAD+. NADH absorbs strongly at 340 nm but NAD+ does not, enabling the reduction of NADH over time to be followed by monitoring the decrease in absorbance at 340 nm (A340). See below for a schematic:
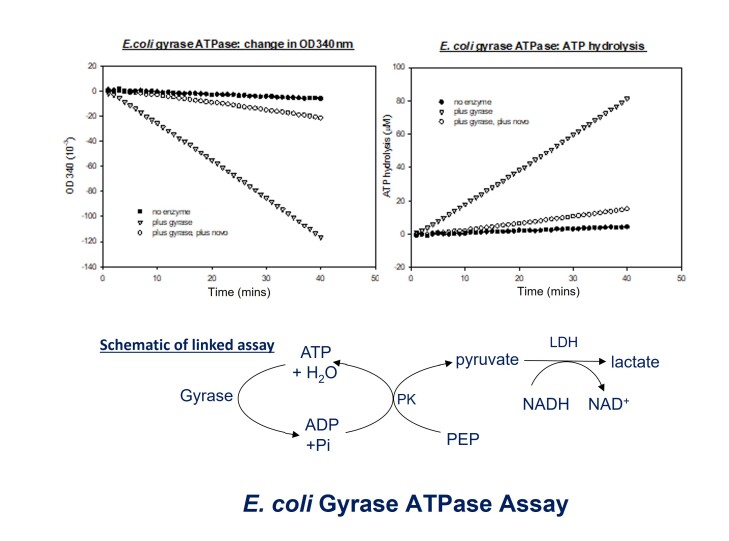
Typical Assays:
Assays should be be carried out under the following conditions at 25°C in 96-well plates:
For determination of DNA-independent ATPase activity:
The N-terminal E. coli Gyr B43 domain is incubated in 1X ATPase Assay Buffer, 800 µM phosphoenolpyruvate, 400 µM NADH, 1% (v/v) PK/LDH (pyruvate kinase-lactate dehydrogenase mixture in 50% (w/v) glycerol, 100 mM KCL, 10 mM HEPES (pH 7.0) ) and 2 mM ATP.
The 43-kDa protein is added to the reaction at a final concentration of 20 µM and allowed to incubate with the compounds in the microtitre plate for 10 minutes before the addition of the ATP. The decrease A340 is then measured over 20 minutes.
For determination of the DNA-dependent ATPase activity:
Gyrase (70 nM) is incubated with linear pBR322 DNA (2.8 nM) under the same conditions described above, except that the NADH concentration is halved. In both cases, a range of drug concentrations can be used to determine the potential inhibitory effects.
Reactions are initiated by the addition of 12 µL ATP (25 mM) giving a final concentration of 2 mM. The decrease in A340 is measured for 20 minutes.
